All Study Data Download
(ABCD & HBCD Studies)
Take me to the File-Based DownloadInstructional Video
Prerequisites
- Globus Connect Personal or Globus Connect Server must be properly installed and configured. For more details, please refer the documentation available here.
- You must be authorized to access the ABCD or HBCD datasets via Lasso Informatics.
How do I download file-based data?
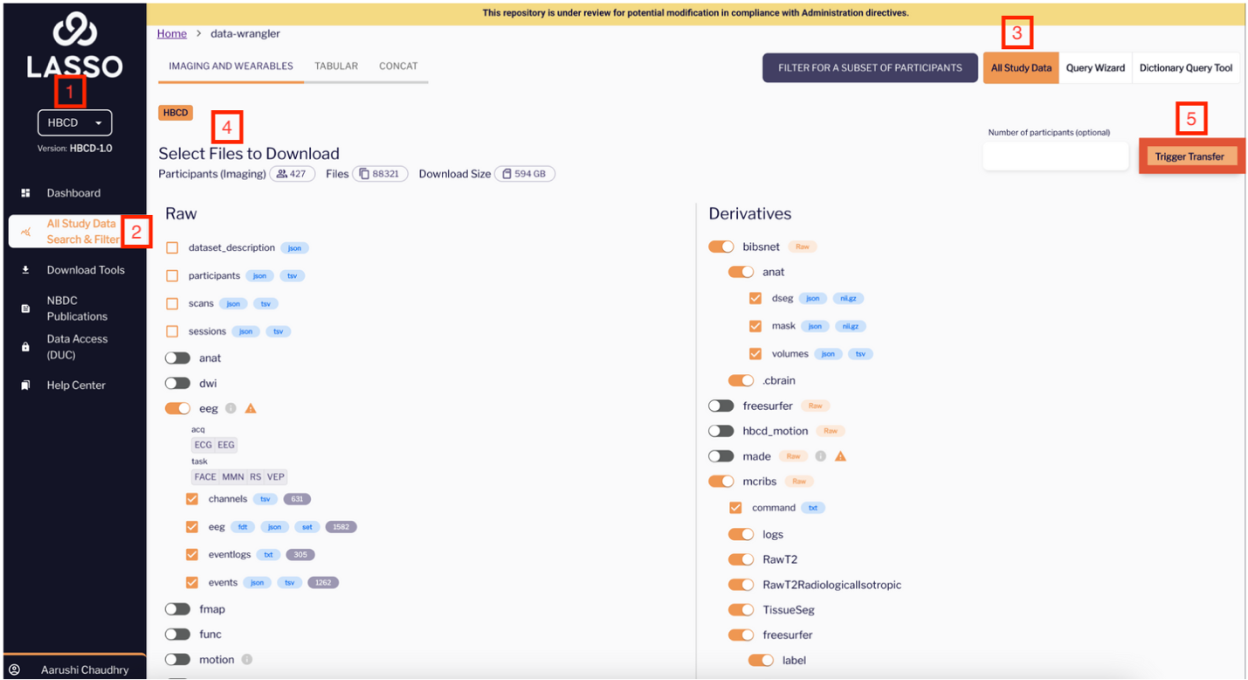
- Choose your study (ABCD or HBCD) from the study selector
- Go to the All Study Data Search & Filter Module
- Click All Study Data in the top right-hand corner
- Select your files of interest using the toggles
- Click Trigger Transfer in the top right corner
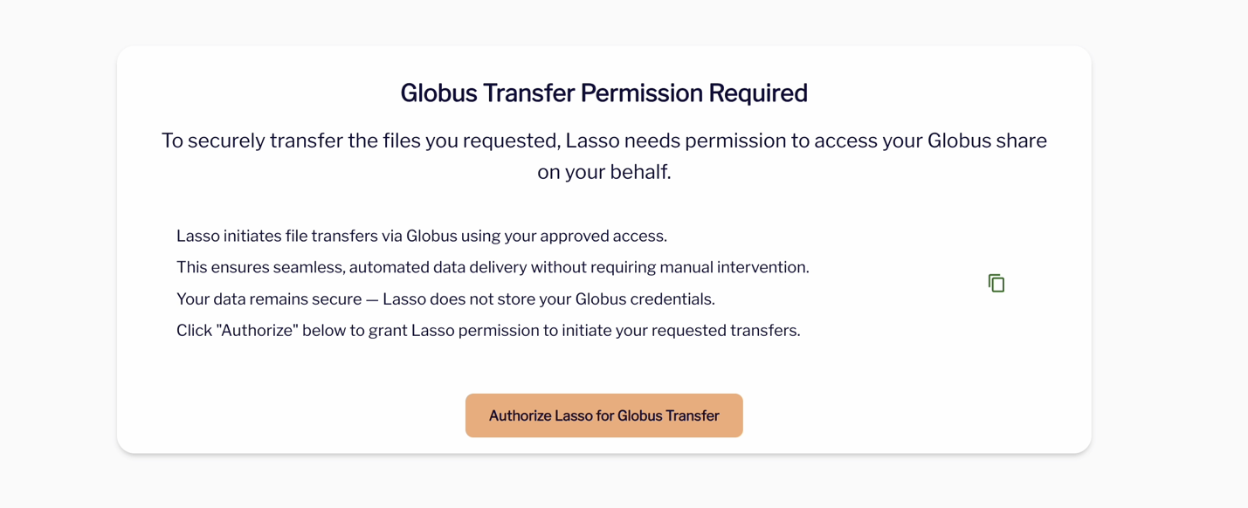
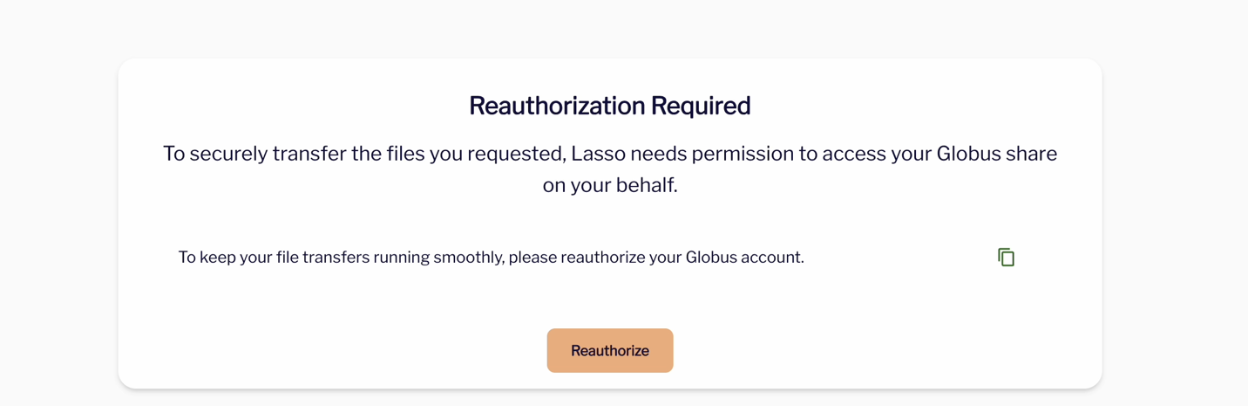
- You will have to authorize or re-authorize Lasso for Globus Transfers.
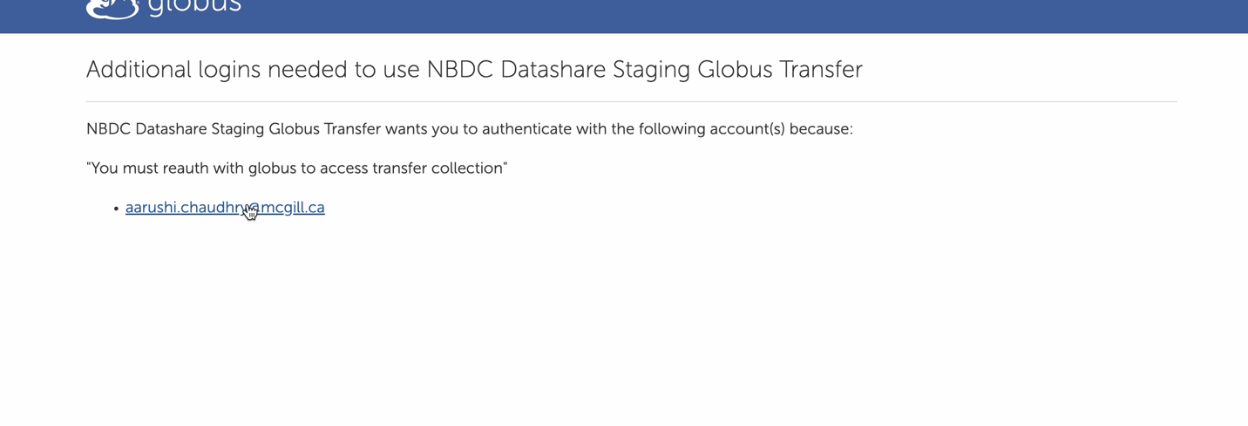
- Please complete the authorization process by clicking on your email and logging in. We recommend using your institutional credentials, or alternatively, your Globus ID.
- Next, you will be redirected to the Globus collections page where you can view your personal collections as well as your Institutional Servers. Double click on your preferred collection container to choose a download destination.
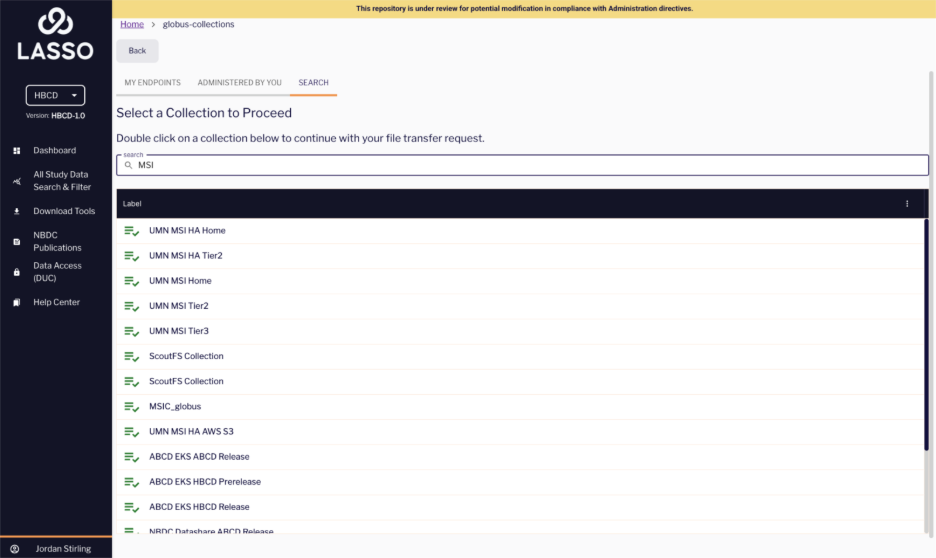
- You can either select an existing directory or folder on your computer or create a new one for your files to be downloaded to.
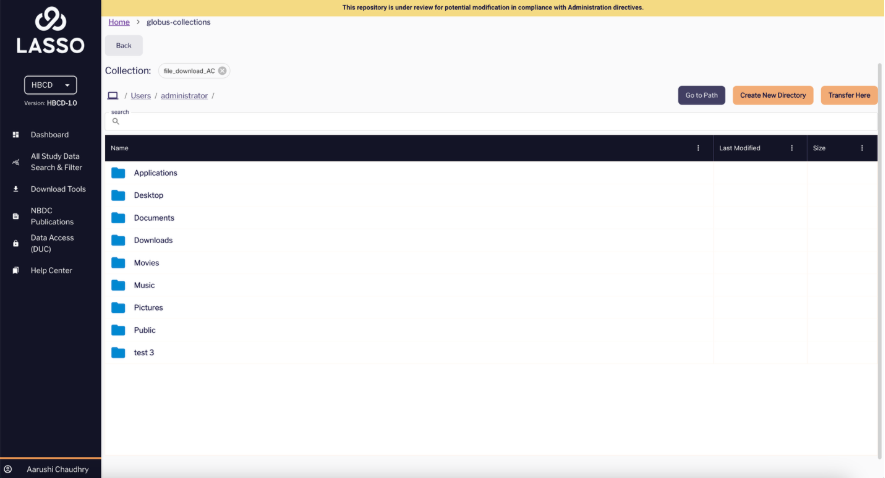
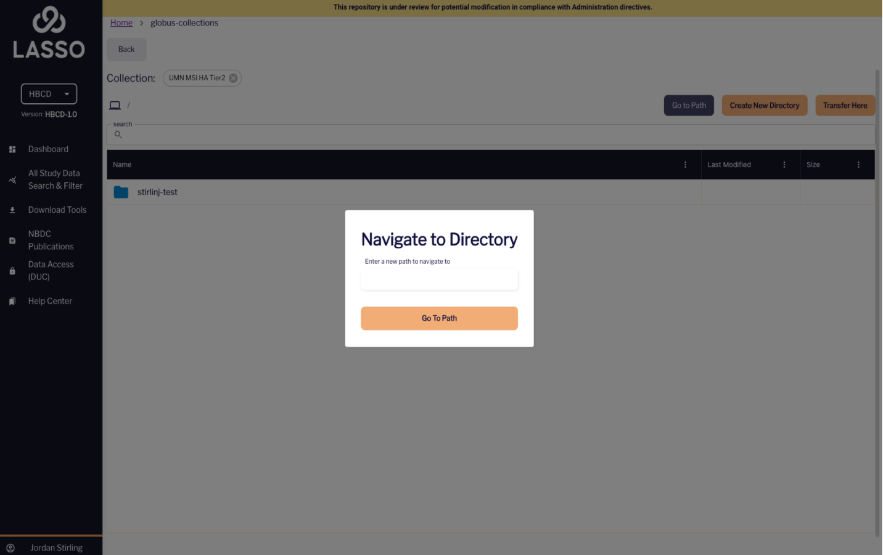
You can also click on the purple “Go to Path” button to search for a specific directory.
- Click on Transfer Here to begin the data transfer process
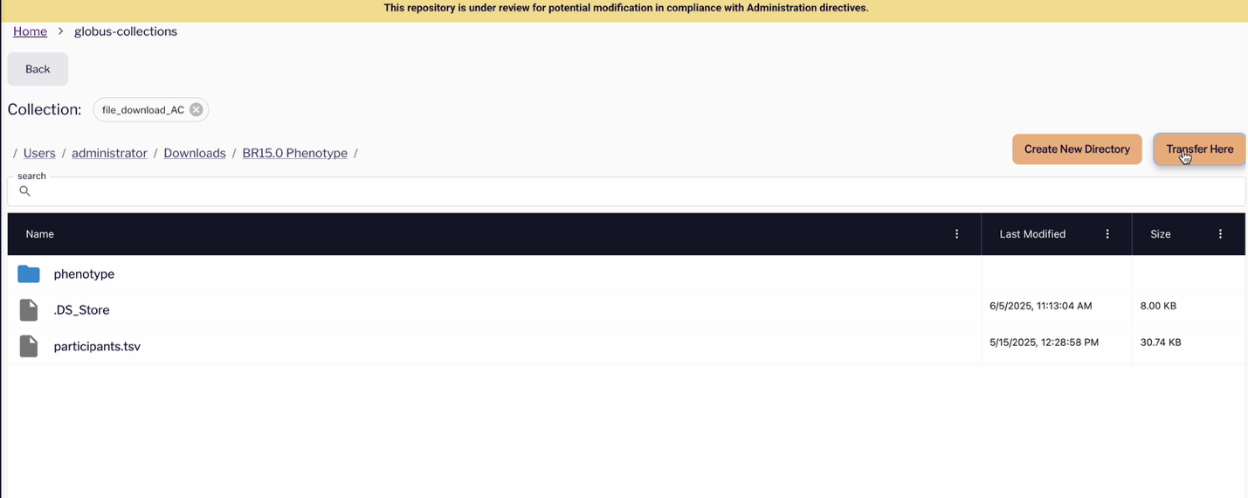
Once the download is complete, your files will be downloaded to the destination specified.
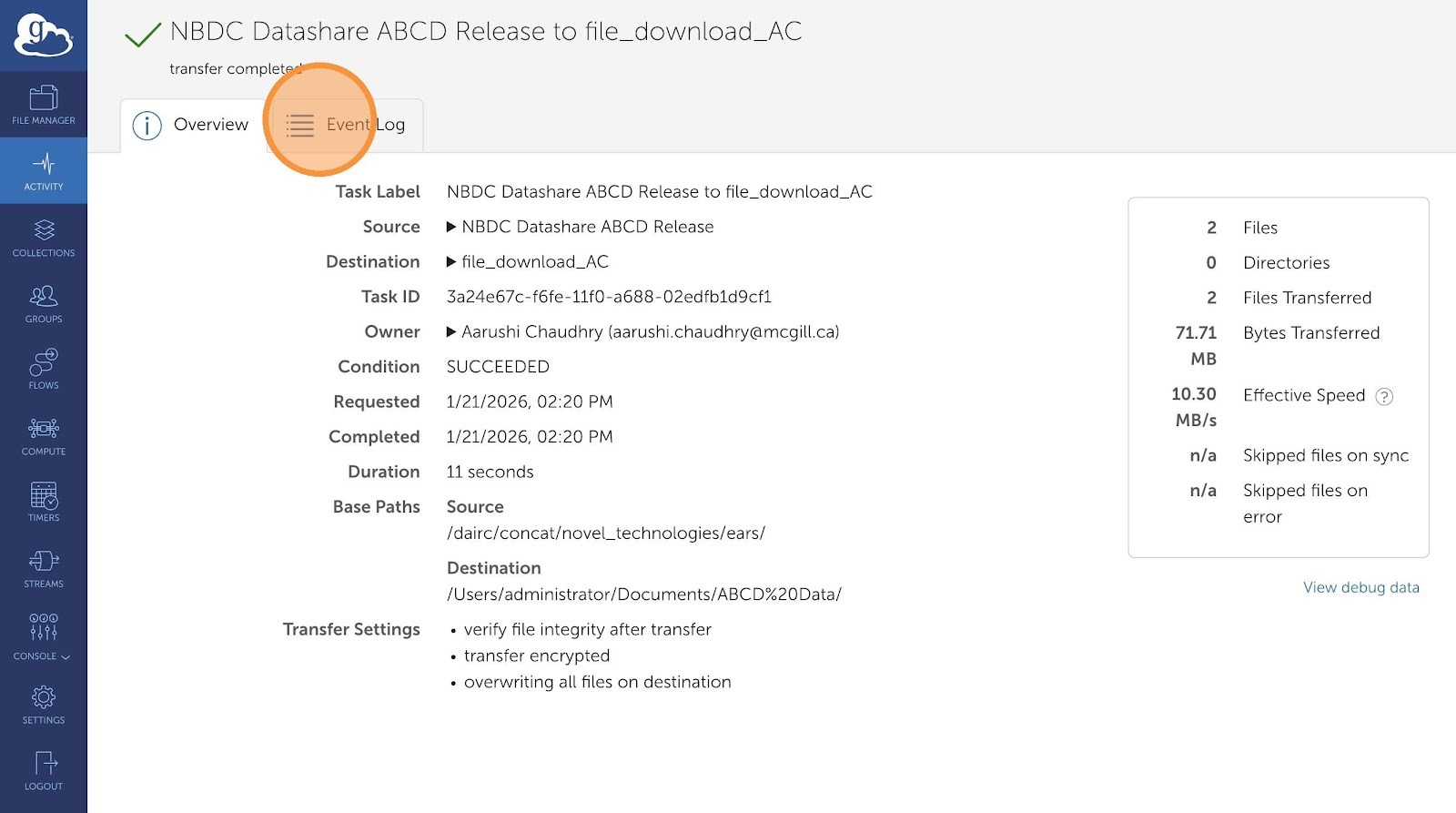
You can check the progress of your data transfer by:
- Clicking on the link provided on this screen
- Or Windows and Mac users can tap on the Globus icon in the systems tray and select “Web: Activity”
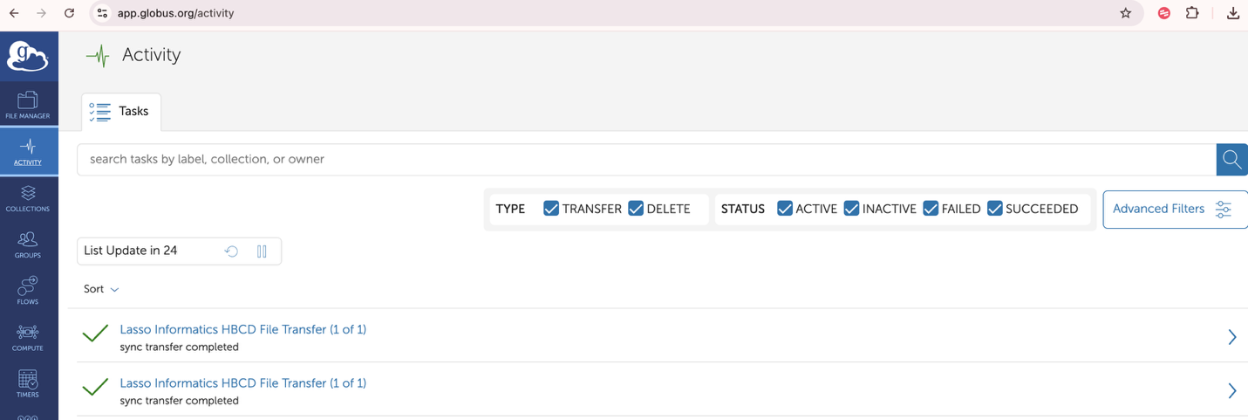
- Visiting globus.org and clicking on activity from the menu on the left side of the application
You will also be sent an email once your data transfer is complete.
Can I download file-based data through the Globus interface?
- Please log in to globus.org using your institutional credentials.
- Navigate to https://app.globus.org/file-manager/collections
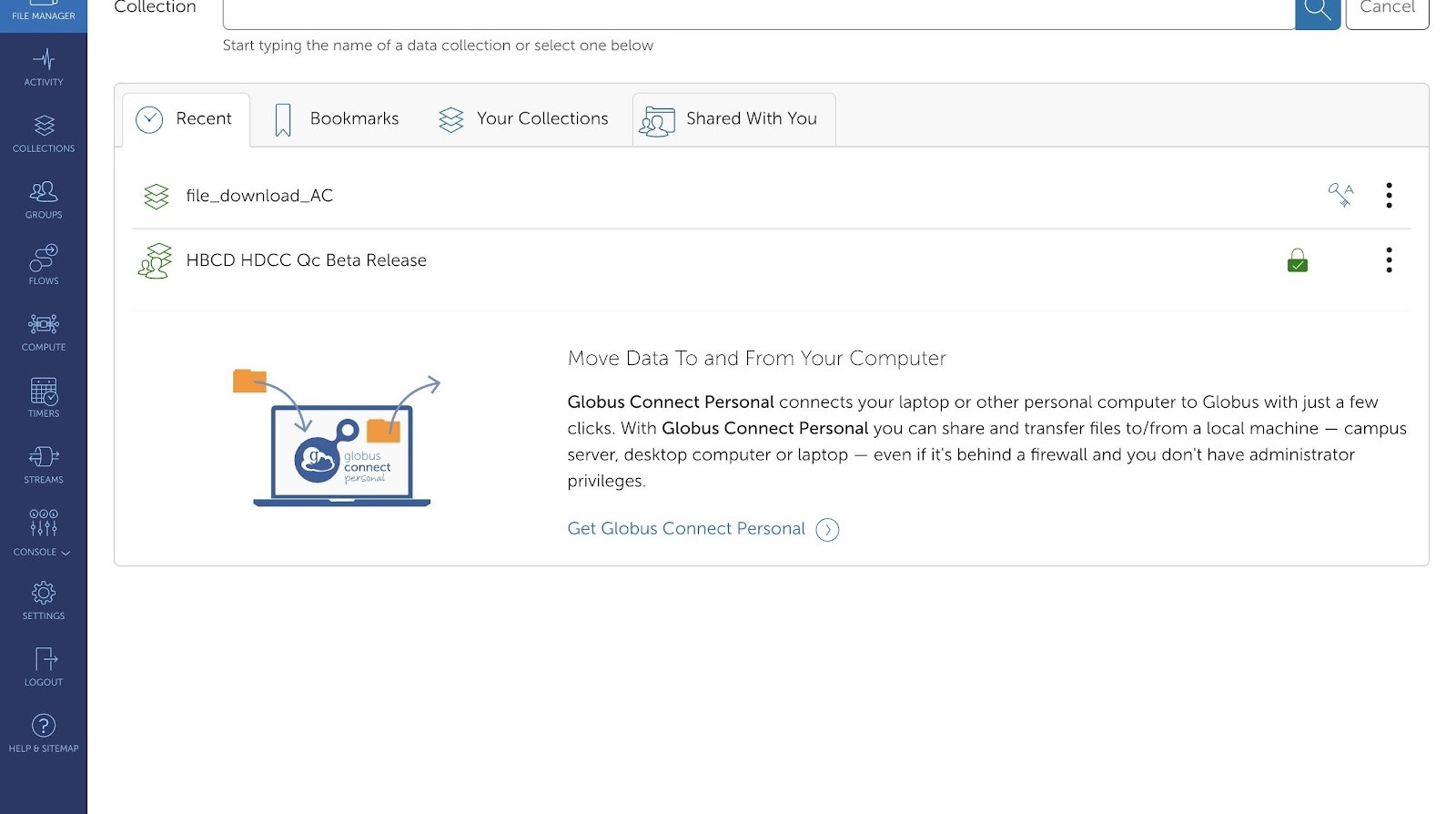
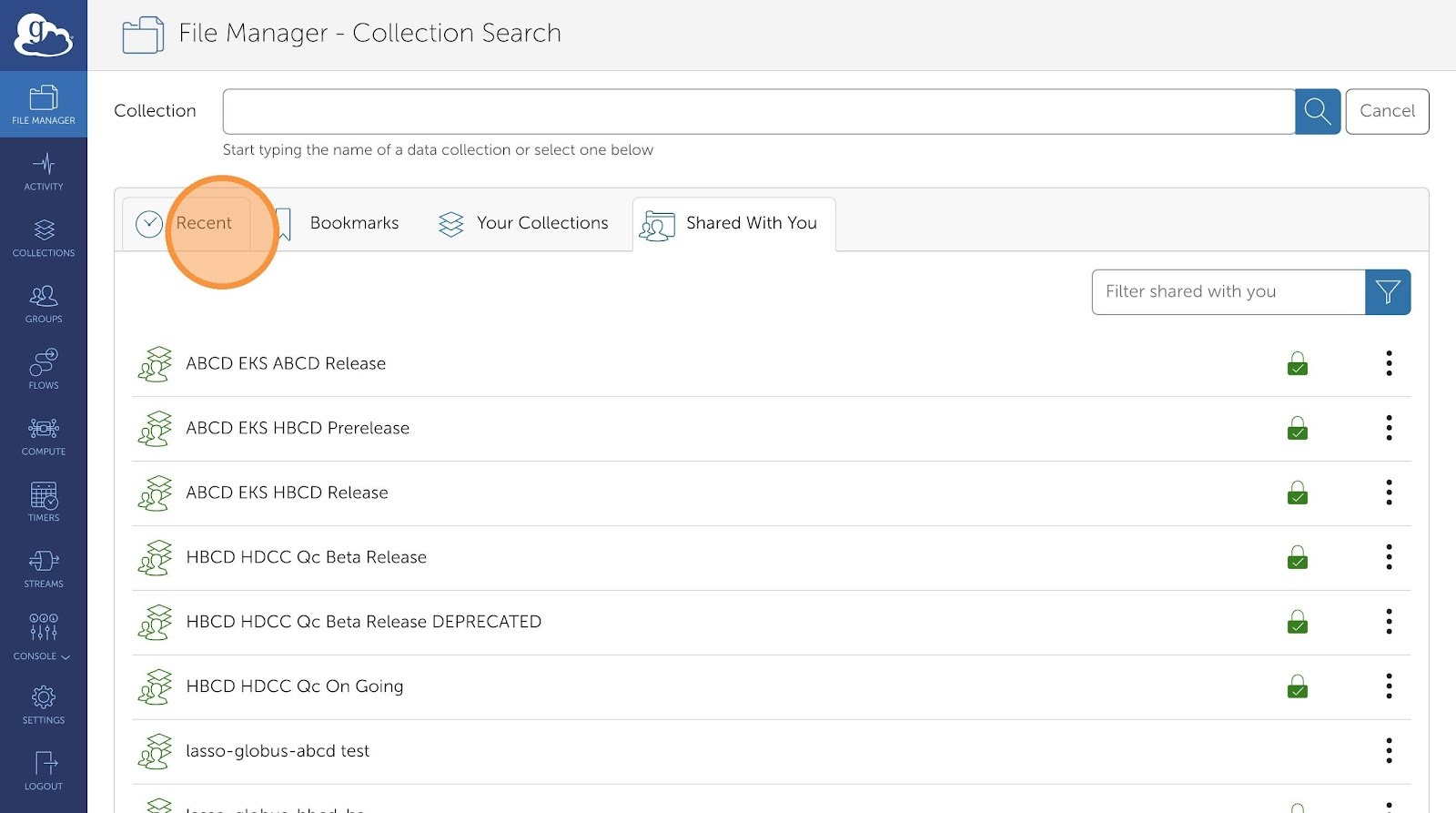
- Click "Shared With You". This will display all the available collections that have been shared with you:
NBDC Datashare ABCD Release
NBDC Datashare HBCD Release
- Click on the collection you want to transfer data from.
- For ABCD, you may see two different data sources, you can check the files available under each of them before starting a transfer.
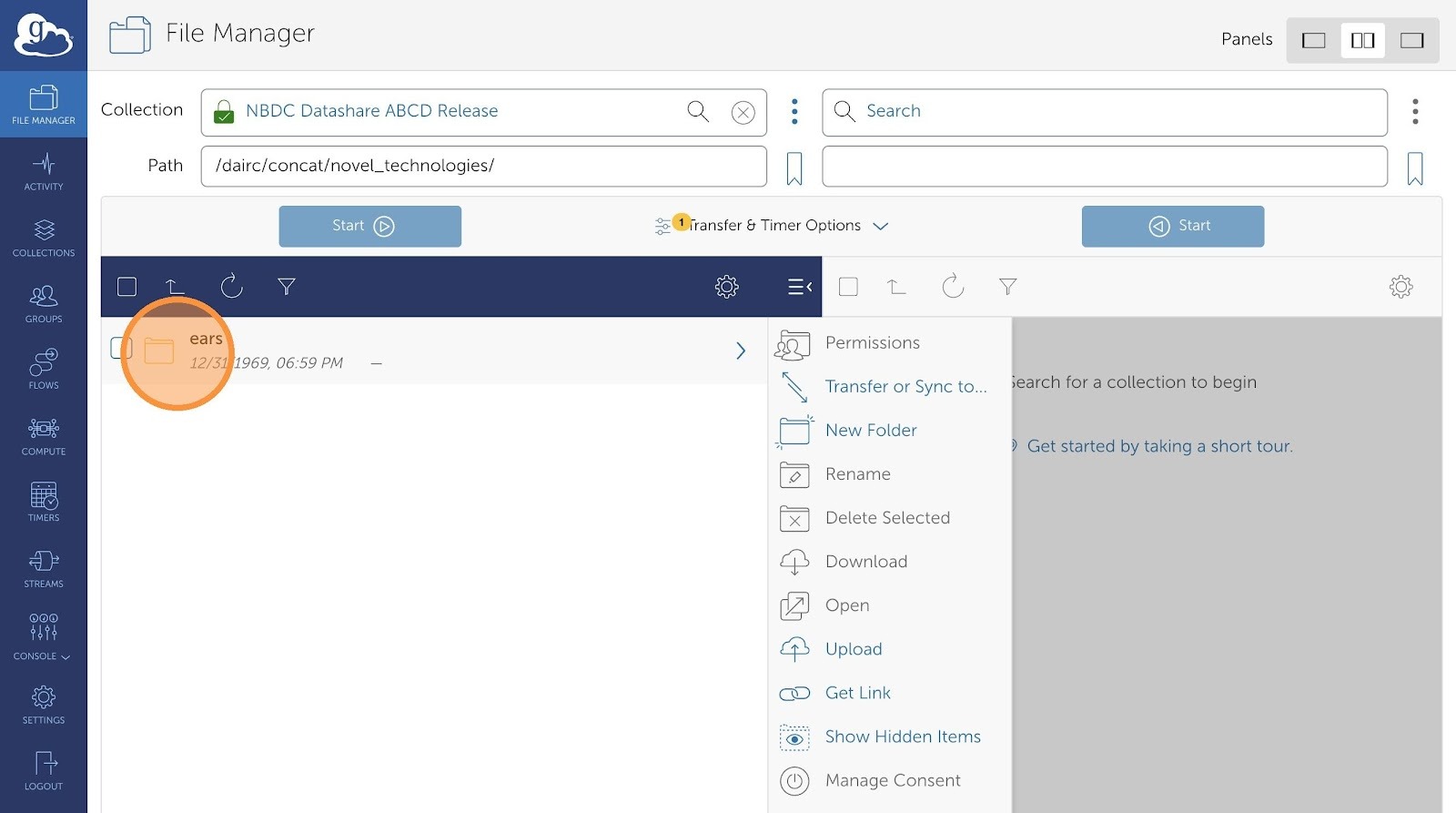
- Select the data type.
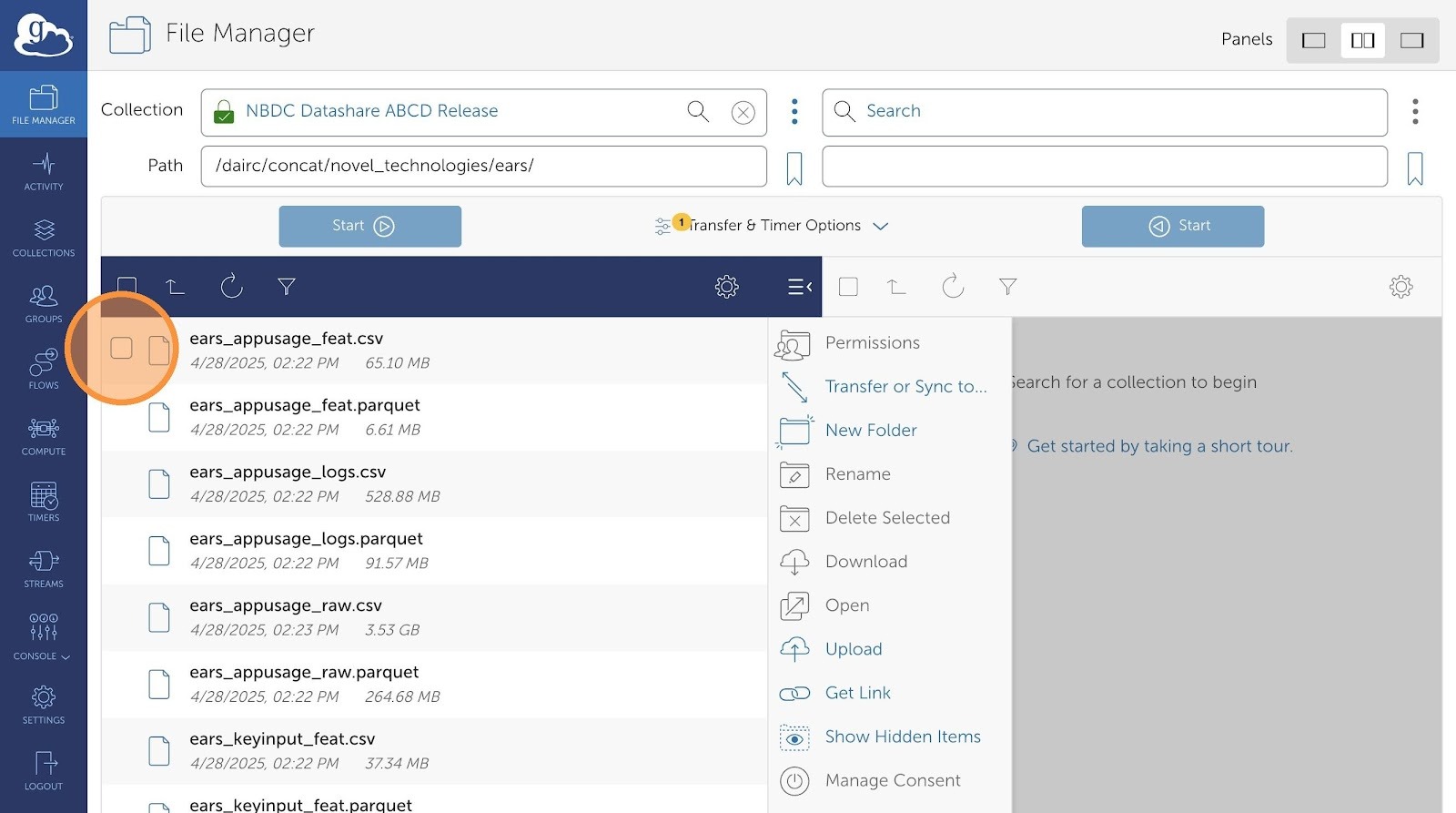
- Now, choose which files you would want to transfer.
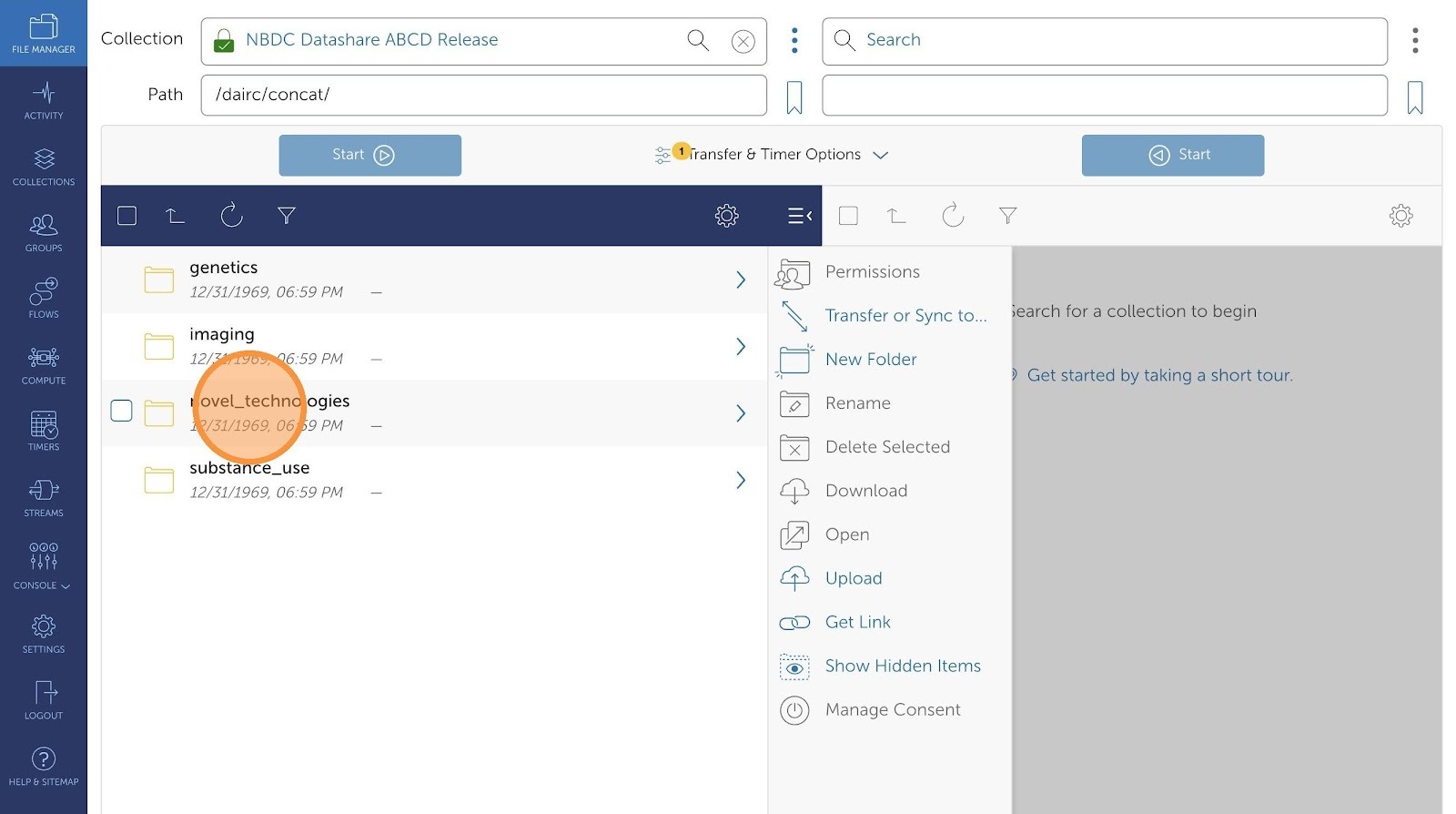
- You can see a list of all available data files to choose from.
- Click the checkboxes to select the files.
- Then, click "Transfer or Sync to..."
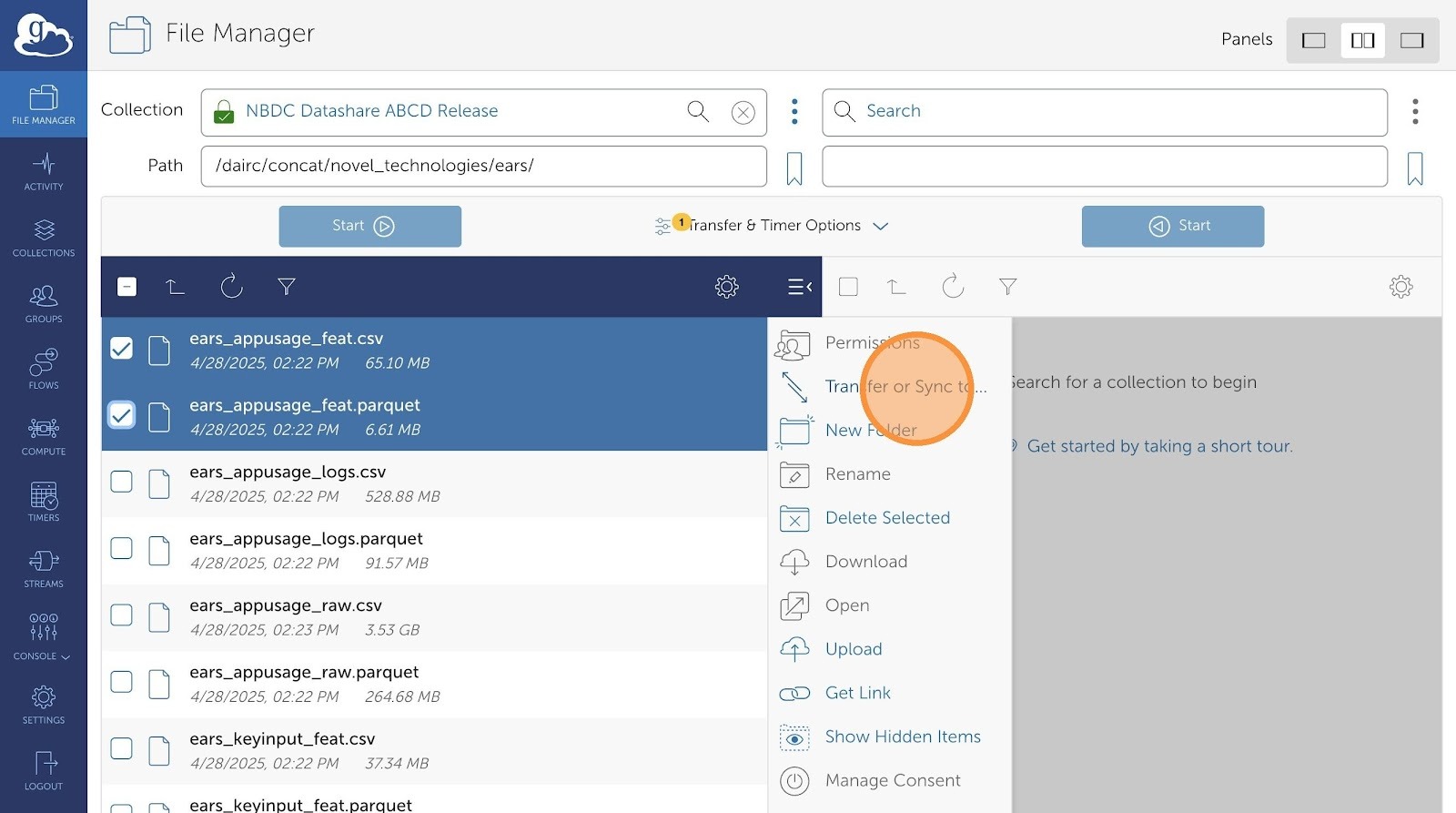
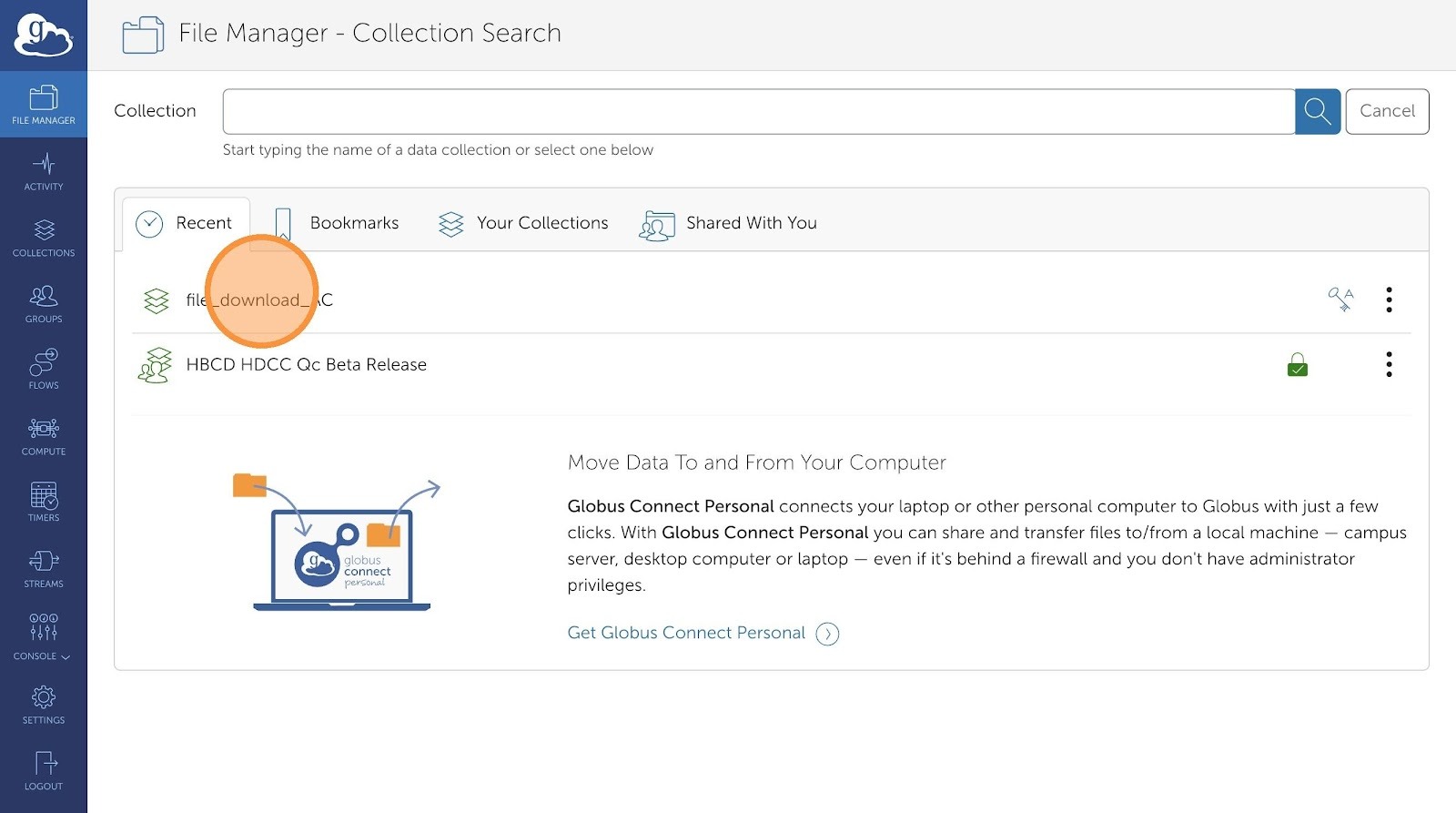
- Click "Search" if you have a destination collection already configured in Globus. Otherwise, type in your file path in the the empty text field next to the bookmark icon (below Search).
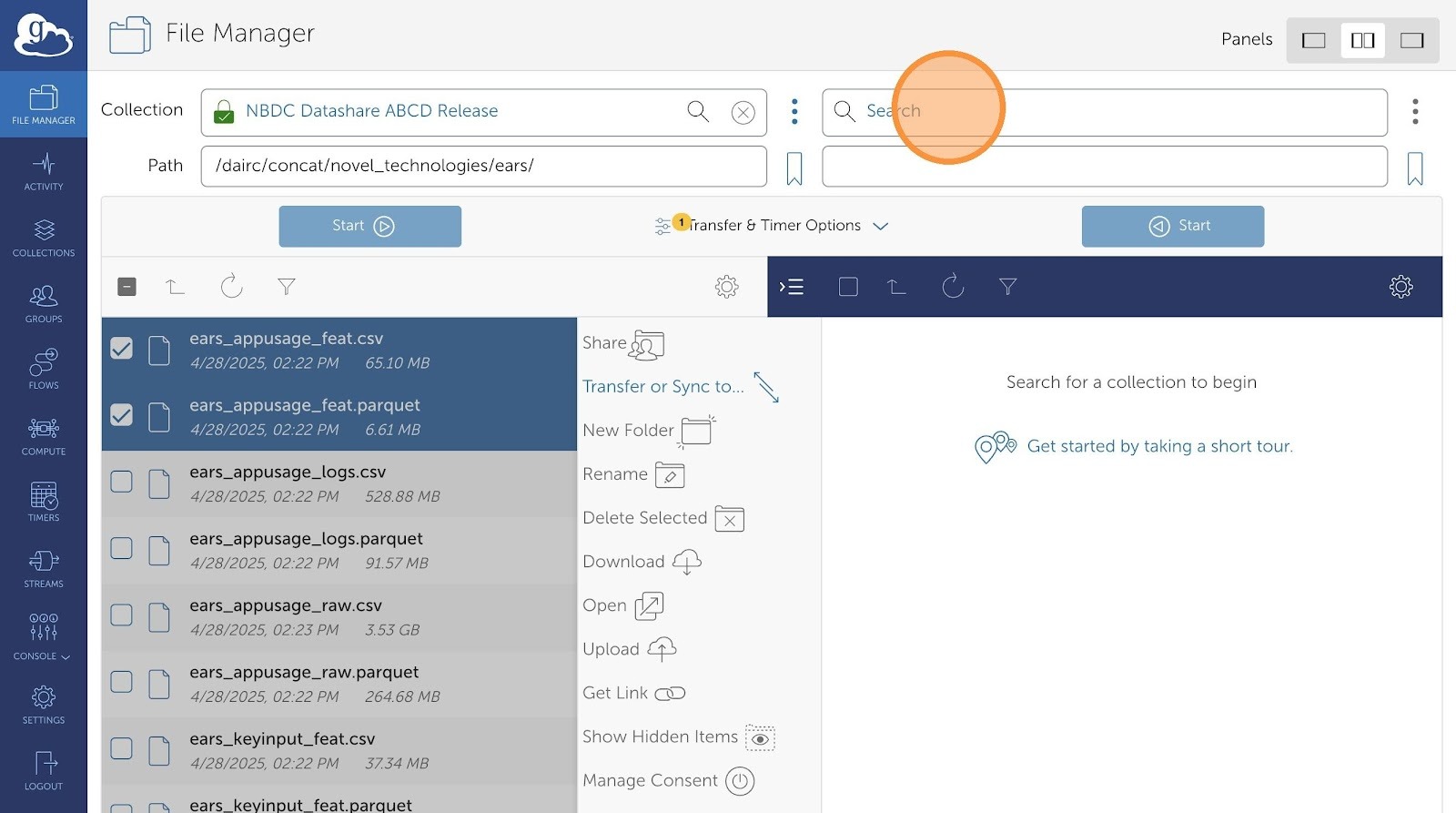
- Search your collection destination.
-
- Now you will see your collection on the right. Select the folder where you'd like to transfer your files.
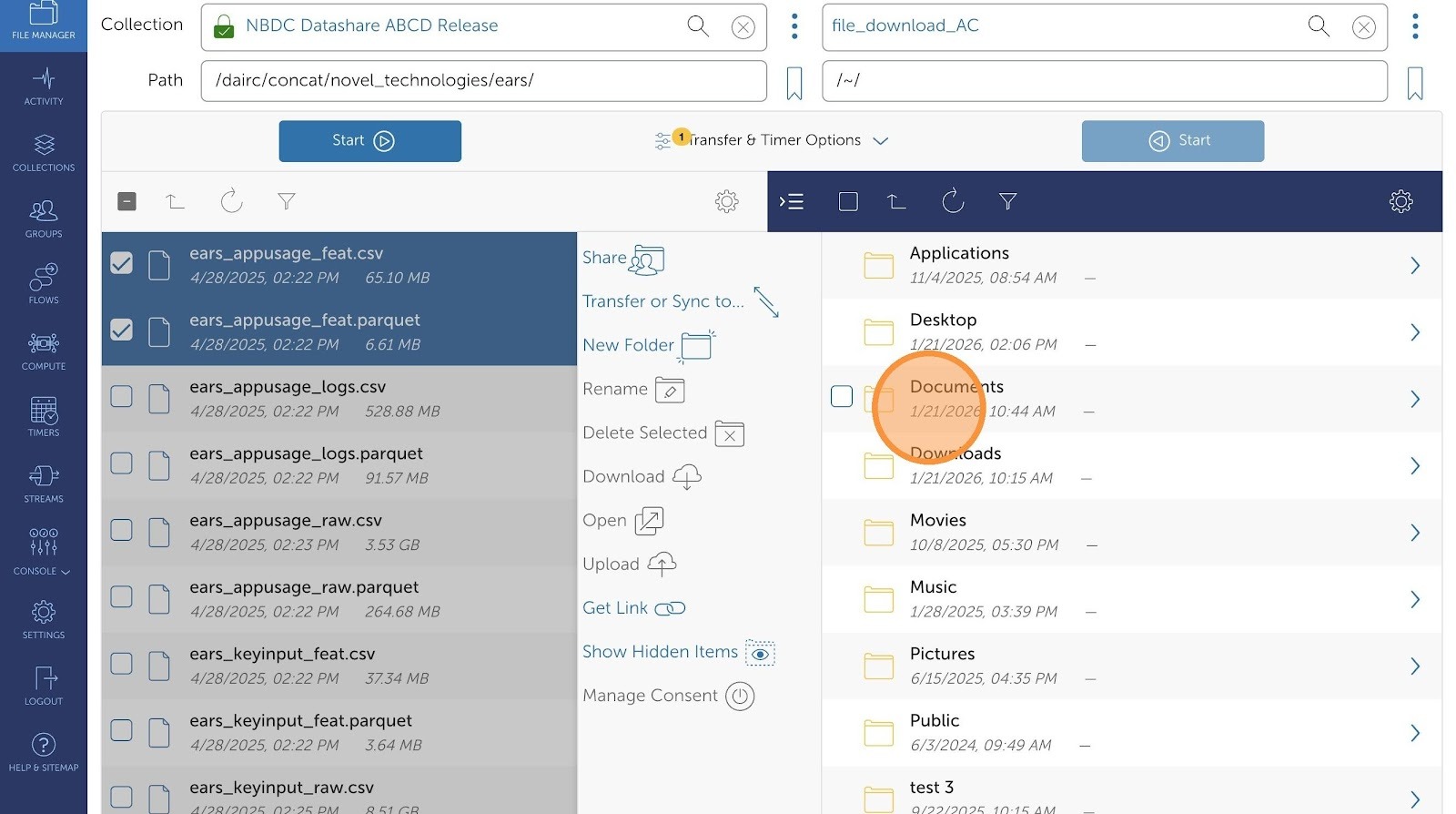
If needed, you can also define various include and exclude rules via the Globus Transfer interface.
For more details, please refer to the filter rules as described in this documentation.
- Filter rules live under Transfer & Timer Options in the middle of the two start buttons.
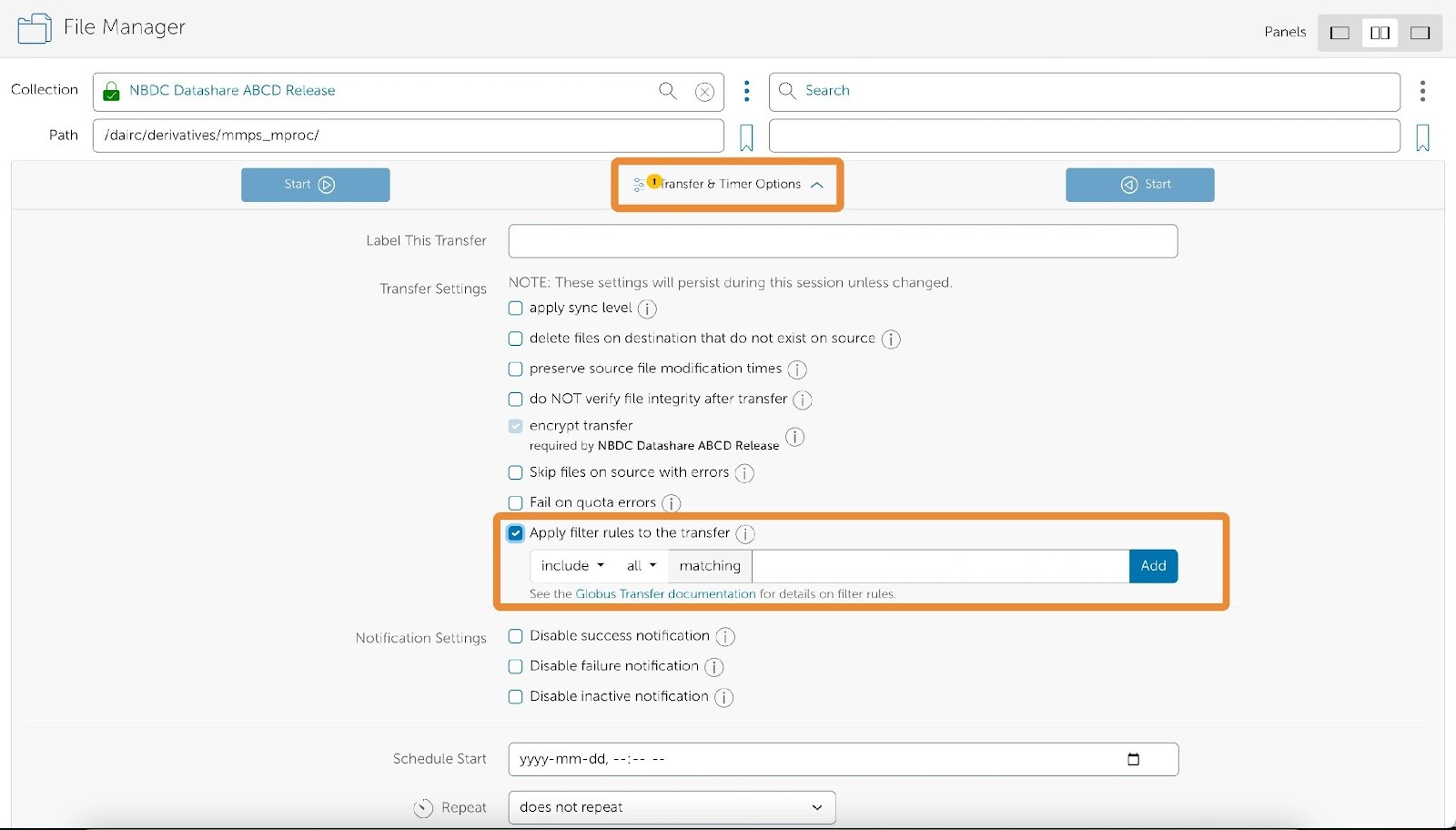
- Now, click start.
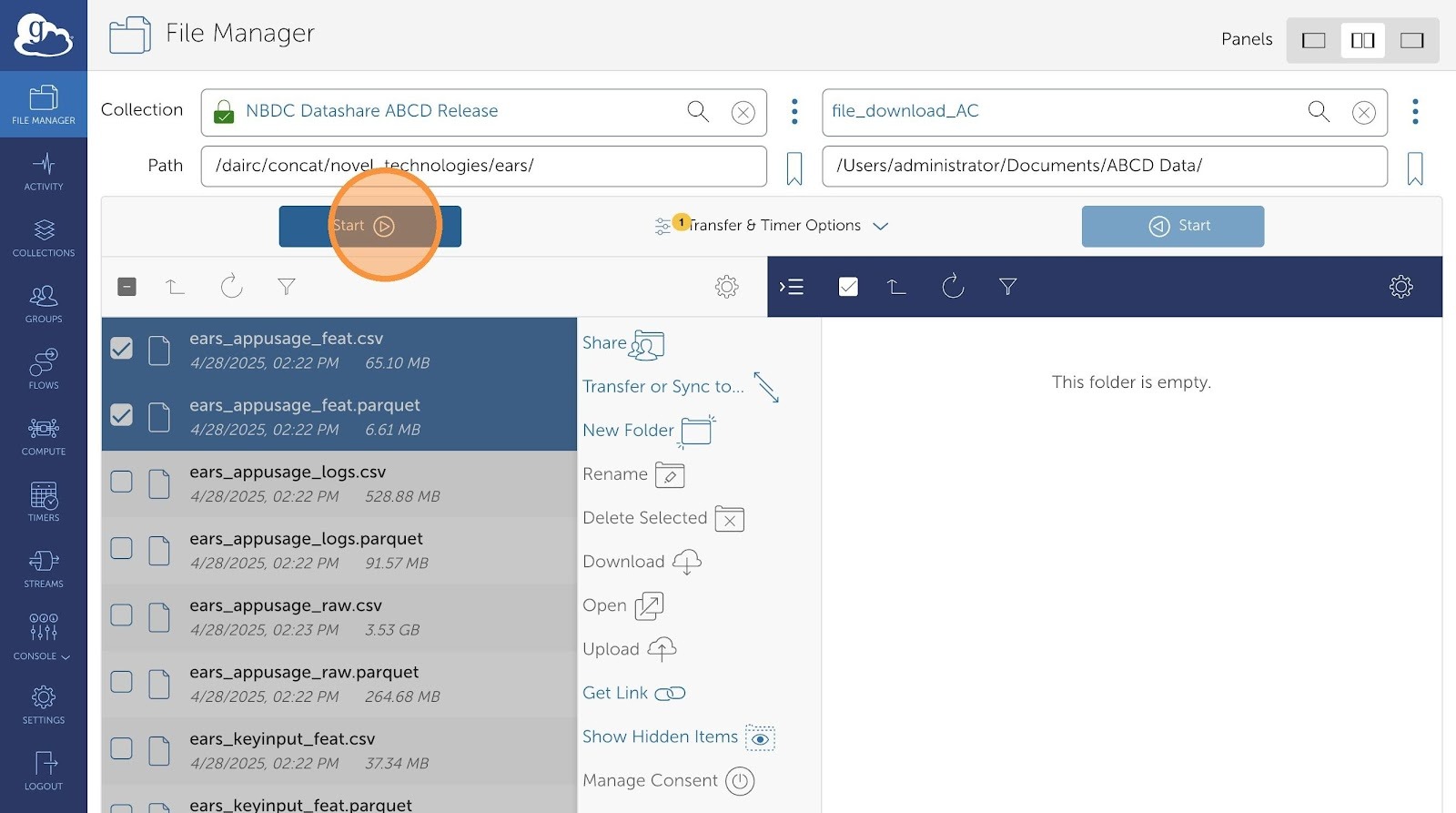
- After your transfer request is submitted, you can check progress in the Activity Tab.
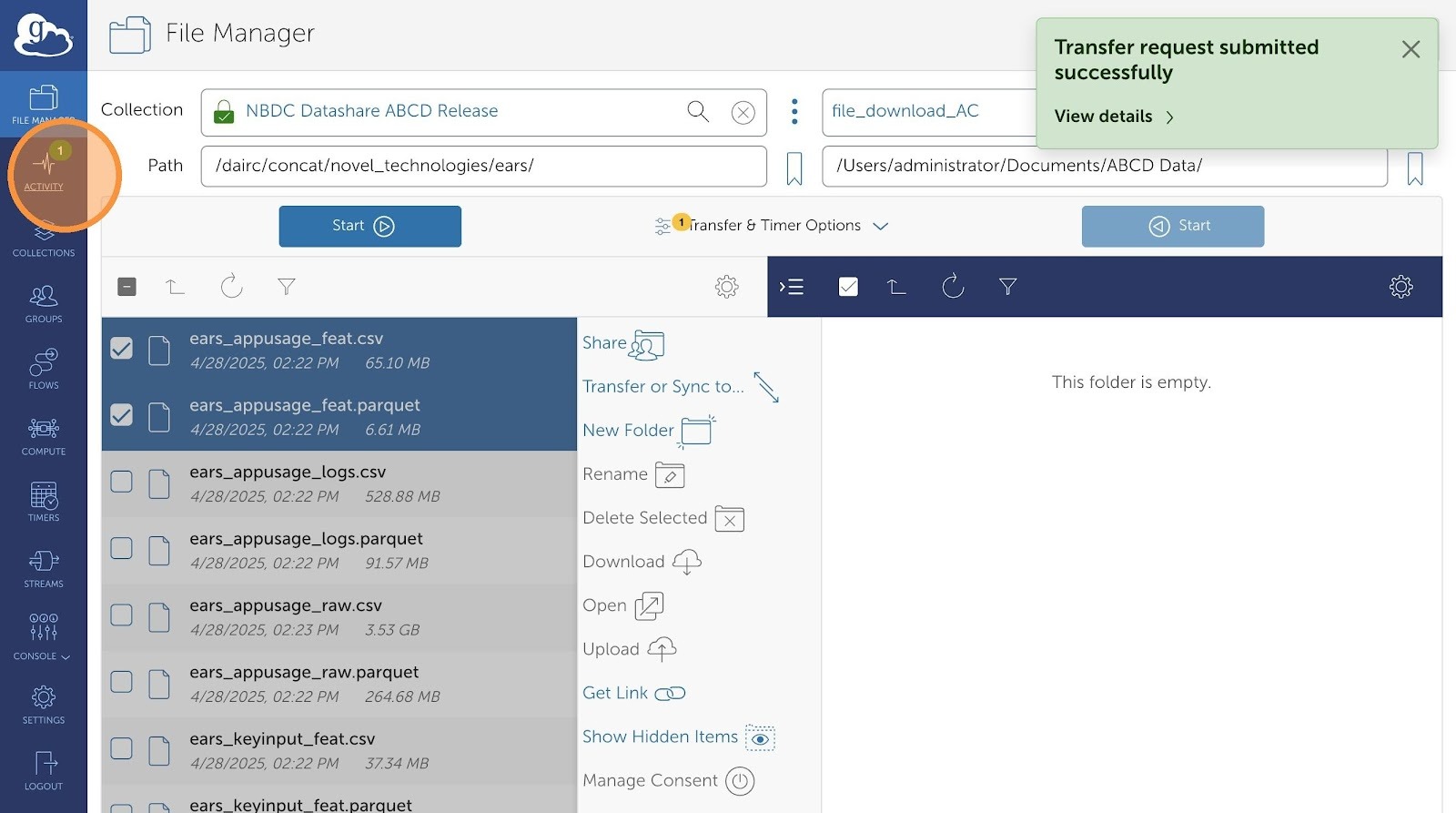
- This dashboard will display history of all your transfer requests. You can view more granular details by clicking on any of the transfers
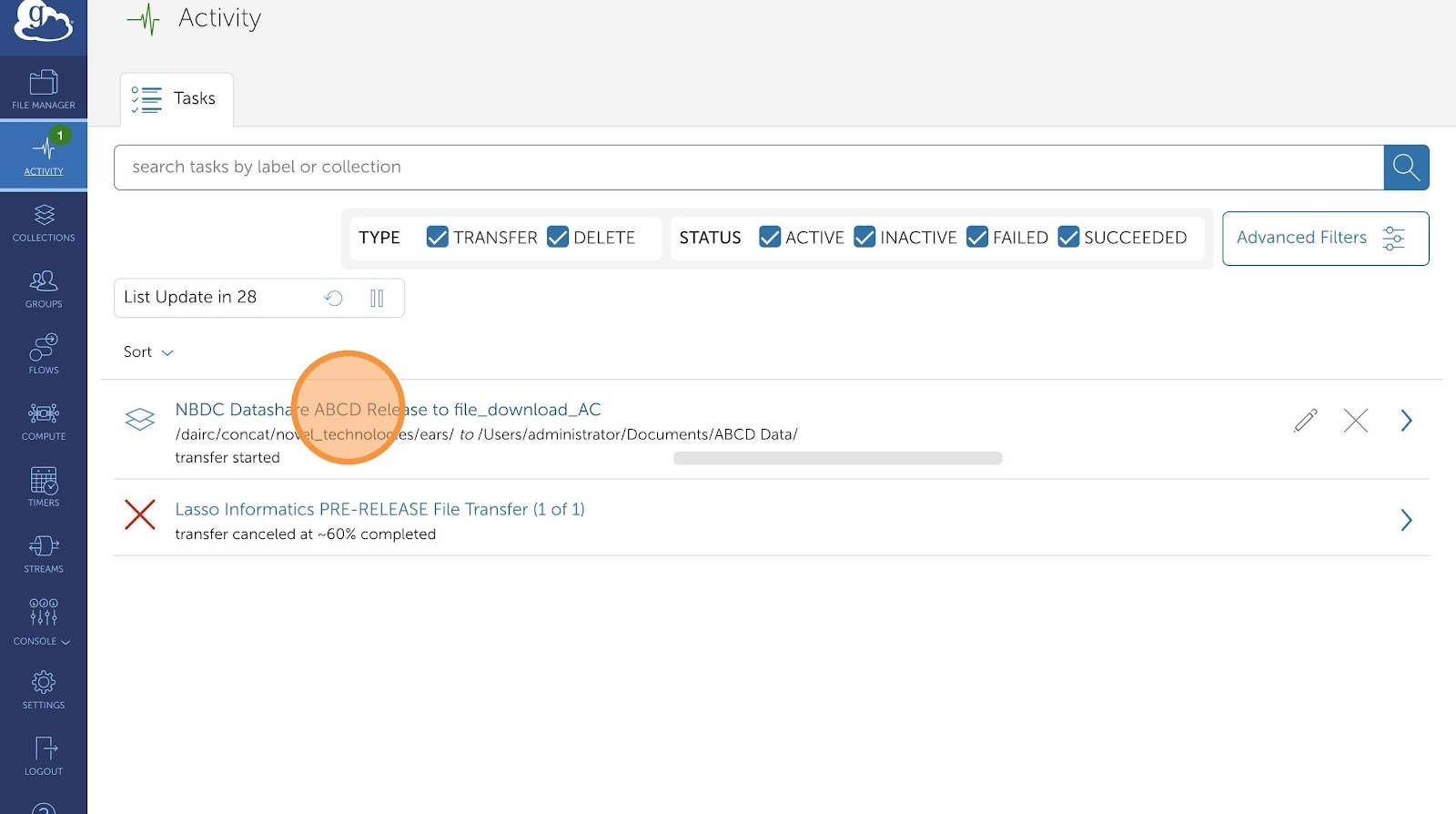
-
Understanding the BIDS Elements in the Lasso file-based query
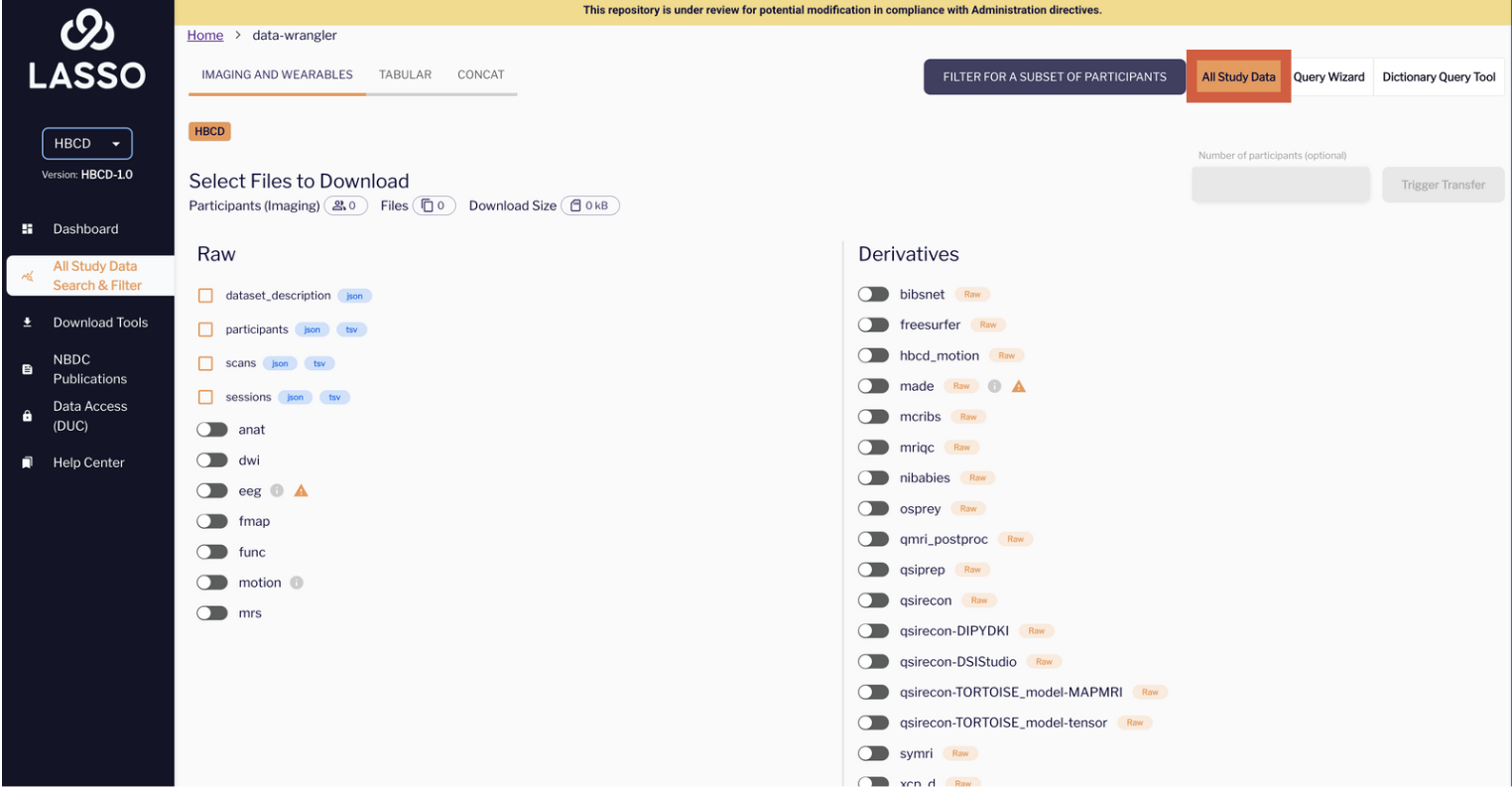
Left Panel: “Raw”
This column lists the core metadata files and imaging modality folders defined by the BIDS standard. In the screenshot above, the items listed below do not have ‘toggles’ but rather check boxes:
- participants.tsv (Tabular)
Lists all participants along with optional metadata such as sex, handedness, and age.
- dataset_description.json (JSON)
Required in every BIDS dataset. Contains critical metadata: study name, version, authors, funding sources, and references.
- sessions.tsv (Tabular)
Provides session-level information for multi-session studies, including visit labels, dates, and site locations.
- scans.tsv (Tabular)
Includes file-specific metadata for each imaging scan—e.g., acquisition time and run number.
- beh/ (Directory)
Stores behavioral task outputs such as events and response data, typically linked to fMRI tasks.
These metadata files serve as the scaffolding for navigating and parsing the BIDS directory structure. They are essential for aligning sessions with scan files and associating imaging data with subject metadata.
Raw Imaging Modalities (Toggles)
Below the files, you see a list of toggle switches labeled:
- anat (anatomical)
- beh (behavioral)
- dwi (diffusion)
- fmap (field maps)
- func (functional MRI)
- mrs (magnetic resonance spectroscopy)
What happens when you toggle these on?
Each toggle enables viewing and selecting corresponding imaging folders under the BIDS /sub-<ID>/ses-<ID>/ hierarchy:
- Anatomical Data (/anat/)
- Includes: T1w, T2w, rec-NORM, rec-NORMALIZED, acq-MPRAGE, acq-SPGR, acq-NORM
- Diffusion Data (/dwi/)
- Includes: dwi, dir-AP, dir-PA, acq-64dirPA, acq-64dirAP
- Functional Data (/func/)
- Includes: task-FEED, task-REST, _events.tsv, _physio.json
- Field Maps (/fmap/)
- Includes: acq-FUNC, dir-PA, dir-AP, _epi.nii.gz
- Behavioral Data (/beh/)
- Includes: blocklog.tsv, events.tsv, tap.tsv, tap.json
- MRS Data (/mrs/)
- Includes: HERCULES_SHORTTE, sub-*/mrs/HERCULES_SHORTTE
Each scan has associated.nii.gz files and .json sidecars for acquisition metadata.
Derivatives Panel
This directory contains derived neuroimaging and behavioral data outputs generated through standardized and study-specific preprocessing pipelines. These derivatives serve as precomputed resources for analysis, quality control, and visualization. Some folders follow the BIDS Derivatives specification, while others use internally defined conventions.
Folder Structure
Each derivative folder is organized by participant (sub-<ID>) and session (ses-<ID>), and may contain:
- Processed NIfTI images
- Surface reconstructions
- Confound regressors
- Motion parameters
- Quality metrics
- Tabular summaries
- JSON metadata (dataset_description.json)
Included Pipelines
ABCD Derivatives
- freesurfer/
Performs cortical surface reconstruction and volumetric segmentation.
Includes subfolders like surf, mri, label, stats, etc.
- mmps_mproc/
ABCD-specific pipeline for structural, diffusion, and functional data preprocessing.
- dataset_description.json
Contains metadata for each pipeline output, adhering to BIDS-derivative guidelines.
HBCD Pipelines
- freesurfer/
Standard surface-based morphometry.
- qsiprep/, qsirecon/
Handles diffusion MRI preprocessing and reconstruction using models from DIPY, DSI Studio, and TORTOISE.
- xcp_d/
Post-fMRIPrep BOLD timeseries processing including nuisance regression and filtering.
- mriqc/
Automates image quality metrics generation.
- qmriqpostproc/, symri/
Supports quantitative MRI postprocessing.
- osprey/, nibabies/, mcribs/
Provides pipelines for infant-optimized processing and spectroscopy.
- hbcd_motion/, madel/, bibsnet/
Includes motion quantification and infant segmentation tools.
- qsigc_*, qsirecon-*, TORTOISE_*
Specialized pipelines for dMRI modeling and reconstructions.
- dataset_description.json
Present in top-level derivative folders for documentation purposes.
Metadata Files
Each pipeline directory typically includes:
- dataset_description.json: Describes the derivative dataset origin and tool versions
- *.tsv: Tabular summaries or metrics per participant/region
- *.json: Metadata describing acquisition, processing parameters, or QC outcomes
Notes and Best Practices
- Not all derivative folders are fully BIDS-compliant; check the dataset_description.json for versioning and schema.
- Some outputs may include additional quality-control images, masks, or intermediate files not suitable for direct statistical analysis.
- When using derived data in publications, cite the appropriate software (e.g., FreeSurfer, FMRIPrep, QSIPrep) and study Date Use Certification (DUC) guidelines.
Right Panel: Source Section (ABCD Only)
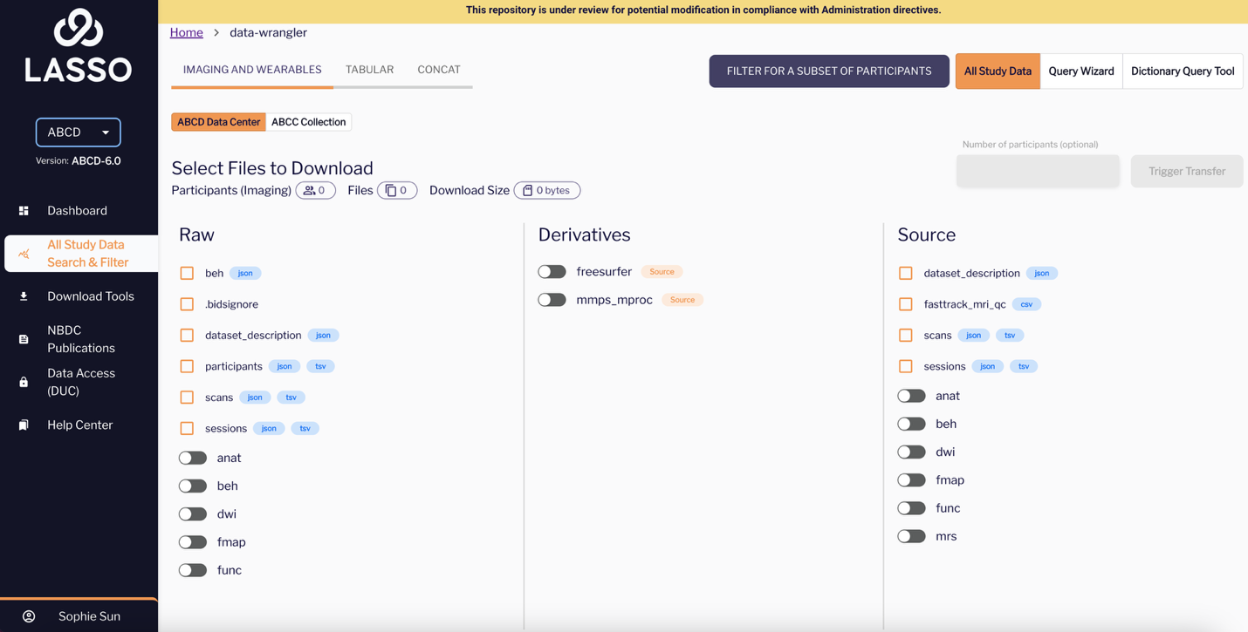
This section lists additional source-level metadata not embedded in the raw folders. These help with study-level quality control or linking files to scan procedures.
Examples:
- fasttrack_mri_qc.csv
Captures imaging quality control flags and metrics across sessions.
- scans.tsv / scans.json
Documents the scans that were executed, including timing information and file-level annotations.
- sessions.tsv / sessions.json
Provides details about study visits, such as scanning site, scanner used, and visit date.
- dataset_description.json
Reiterated here for clarity—offers an overview of the entire study including its structure and metadata.

